Removing Dummy Metal Cells
Large GDSII files are made even larger when prepared for maskmaking by the addition of dummy fill metal. The fill metal is needed to produce a uniform metal density on the various conductor layers - if these layers are not uniformly dense, then the polishing and CMP processes will not be uniform either.
This dummy metal fill can make the already large file much much larger and makes it more difficult to open for viewing or to perform operations that essentially do not need or use the dummy metal fill.
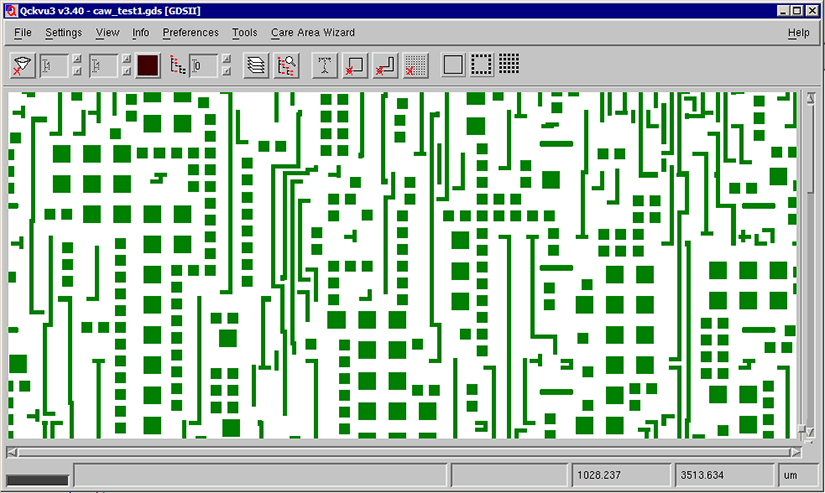
Figure 1: The pattern of squares represents dummy fill. For this layer, there are more fill entities than actual circuit traces.
Our objective is to remove the dummy metal fill without affecting the rest of the design data. The result will be a new GDSII file that is considerably smaller, opens faster in our viewer and can be processed (for example for net tracing or care area creation) much faster.
How Dummy Fill is Identified
Since the dummy fill metal is generally added as a postprocessing step after the layout is completed, it is generally found in cells that are distinct from the basic chip layout. If we are lucky, all the dummy metal is placed under its own hierarchy but this is not always the case. Hopefully the cell names used to contain dummy metal can be isolated using one or more wildcards.
Our approach will be as follows:
- open the original GDSII file to determine the names used for cells containing dummy file
- generate a list of the names
- run GDSFILT and pass it the list of cell names to "knock out."
Example 1 - caw_test1.gds
Our first example is a 1.5GB chip file called caw_test1.gds. When loaded into Qckvu3 the program uses 1.9GB resident memory (the db is loaded into memory and not left on disk.) We are going to remove the dummy metal cells and see how much the file is reduced in size.
Results from the top command (RH4)
PID USER PR NI VIRT RES SHR S %CPU %MEM TIME+ COMMAND 28414 stevedb 15 0 2096m 1.9g 11m S 0.0 57.1 0:57.53 qckvu3.exe
Step 1 - Scan the GDSII File to Get Cell List
We will use the gscan utility to scan the file and to output a list of cell names. The syntax to do this is shown below:
$ gscan caw_test1.gds -sh -sea -elmcnt -dtp -o:caw_test1.txt
About 60 seconds later, we get the output, caw_test1.txt.
A small extract of the file is shown below:
STRUCTURE_LIST 1) DM7_L_DMx 2) RF1SHD_64X28DECODER_DMx 3) RF1SHD_64X28CK2_DMx 4) RF1SHD_64X28HXPS_DMx 5) RF1SHD_64X28M24BOT_DMx 6) RF1SHD_64X28_DMx 7) RF2SH_128X20CTRL_SA_R_DMx 8) RF2SH_128X20_DMx 9) RF2SH_32X22CTRL_SA_R_DMx 10) RF2SH_32X22_DMx 11) RF2SH_256X19BCOL_M_DNMY_DMx 12) RF2SH_256X19CTRL_SA_R_DMx 13) RF2SH_256X19HXP24_DMx 14) RF2SH_256X19HXP24R_DMx 15) RF2SH_256X19HYP24_DMx 16) DM2_S_DMx 17) DM1_S_DMx 18) RF2SH_256X19_DMx 19) RF2SH_512X18BCOL_M_DNMY_DMx 20) RF2SH_512X18CTRL_SA_R_DMx 21) RF2SH_512X18_DMx 22) RA1SHDbit_1024X32HBFX_DMx . . . 2366) RA1SHD_800X28VIANOMX32B 2367) RA1SHD_800X28VIA_CD_DRSTUB 2368) RA1SHD_800X28VIA_CD_DWSTUB 2369) RA1SHD_800X28CDXX 2370) RA1SHD_800X28CDX 2371) RA1SHD_800X28MUX_CD_ODD 2372) RA1SHD_800X28SA8 2373) RA1SHD_800X28VIAIOSG 2374) RA1SHD_800X28IO8 2375) RA1SHD_800X28VIA_IO_WESTUB
Examination of this output shows us that there are many structures that look like metal fill. These structures names end with _DMx, indicating Dummy Metal X (hatch or fill). In fact, there are almost 400 cells that end with _DMx which contain only dummy metal. We are done with this step as it is used merely to understand how we need to identify the dummy fill cell names. If you already knew the naming convention for dummy fill cells, you could skip this step entirely.
Step 2 Creating a Regular Expression for the Remove List
GDSFILT has a -remove command line option that takes as an argument a file. The file can contain either a list of cell names to remove or it can use a wildcard entry. In our case, since we would have to create a list of 390 cell names it is easier, if we understand regular expressions, to use a wildcard.
The expression that will match our dummy fill cells is:
^.*DMx$
Short Explanation of this Regular Expression
"^" indicates the beginning the string to be matched
".*" matches zero or more occurences of any character
basically we don't care what happens in the front of
the structure name string.
"DMx$" the last three characters of the string must match DMx.
this does our filtering work for us.
Step 3 - running GDSFILT with -remove
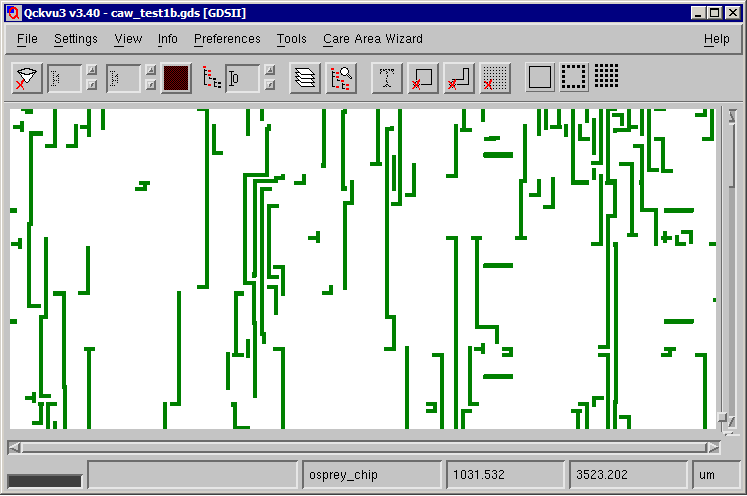
We now run the file, caw_test1.gds through GDSFILT using the -remove command line argument which will remove any cells in the resulting output file that match our regular expression. The output file should be a lot smaller and easier to handle.
$ gdsfilt caw_test1.gds caw_test1b.gds = -remove remove_list.txt where gdsfilt name of the filtering exectuable caw_test1.gds our input file caw_test1b.gds the output file to create = use the top structure -remove the remove command line argument remove_list.txt a text file containing a single line: ^.*DMx$
When loaded into Qckvu3 the new file has a smaller memory footprint: 1.2 GB resident vs. 1.9GB resident.
PID USER PR NI VIRT RES SHR S %CPU %MEM TIME+ COMMAND 30657 stevedb 15 0 1353m 1.2g 11m S 0.0 35.2 0:57.28 qckvu3.exe
Eliminating Via Cells
We also noticed that this same design had a lot of cells dedicated to holding arrays of vias. For Care Area definitions and computations, it is certainly reasonable to eliminate these cells since they do not affect the extents of any cell (vias are always embedded inside of the metal traces) and we don't use them to compute alignment targets.
The via cell names all start with the partial string:
$$via which means we can easily select them with a regular expression.
The expression is:
^\$\$via.*$ where ^ beginning of string \$ match the $ character (escaped with \ because $ is a control character) \$ match the $ character (escaped with \ because $ is a control character) via match the character string via .* any number of characters thereafter $ end of the sting
So our remove_list.txt file now has two lines:
^.*DMx$ ^\$\$via.*$
The first line knocks out any cells ending in DMx and the second line knocks out any cells starting with $$via.
Our gdsfilt command line is:
$ gdsfilt caw_test1.gds caw_test1c.gds = -remove remove_list.txt
and when we open the GDSII file caw_test1c.gds in Qckvu3 we see that the memory footprint has gotten even smaller:
PID USER PR NI VIRT RES SHR S %CPU %MEM TIME+ COMMAND 31367 stevedb 15 0 1112m 945m 11m S 0.0 27.9 0:43.95 qckvu3.exe
